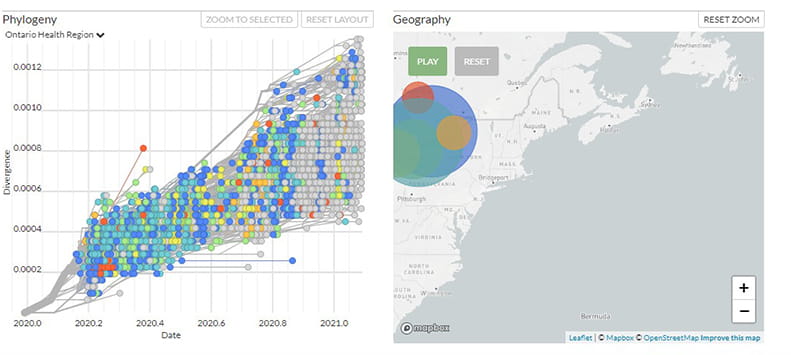
Phylogenetic Analysis of SARS-CoV-2 in Ontario
Data in the tool can be filtered by:
- Ontario Health Region
- Pango lineages, including variants of concern lineages
- age group
- sex
- genotype
- country
- Clades: Nextstrain and GISAID
View Phylogenetic Analysis of SARS-CoV-2 in Ontario
The tool is updated Thursday between 4 and 6 p.m. ET and may be unavailable at this time. View the Technical Notes for more information on what is included in the tool.
Nextstrain
This interactive tool uses the open source platform Nextstrain, which provides powerful analytic and visualization tools to support genomic analysis of pathogens like SARS-CoV-2.
To learn more about Nextstrain or support using and interpreting data in the tool, visit the Nextstrain documentation webpage for a variety of resources, tutorials and user-guides.
PHO Microbiology Rounds: Welcome to the Machine (Learning): Leveraging Artificial Intelligence for Stool Parasite Detection
This PHO Microbiology Rounds will explore three such advents of AI in clinical parasitology that are in use or in development.
Don’t have a MyPHO account? Register Now